Estimating Subgroup Heterogeneity in Difference-in-differences
Kyle Butts and Brantly Callaway
This post addresses a common question we have been asked regarding “I have used the Callaway and Sant’Anna” estimator to estimate some average treatment effect for a population, but want to see if the treatment effect varies along different dimensions. This could be for units of different groups like gender, race, or college-degree (discrete ) or for binned/discretized continuous variables like age-bins (discretized ). The goal is to estimate the group-time ATT for subgroups defined by a specific for a given value of .
One way you can think about the Callaway and Sant’Anna approach is that it provides a sort of rubric for how to estimate aggregate parameters: if you can devise a way to estimate an effect for a , then the aggregating up the “ building blocks” to say an event-study (dynamic) or an overall treatment effect is “standard”.
Brantly has even invested in creating a package,
pte, which allows for a
“plug-in-play” approach to this. In this package, Brant does all the
wiring and you only have to tell it how to estimate the treatment effect
for a given dataset.
In this post, we will disucss estimating a subgroup-specific
and then show how to code this in the pte package. A complete example
is available in this gist.
Subgroup-
The basic set-up follows Callaway and Sant’Anna where you observe units treated at different points in time, denoted by . You are willing to impose that conditional on a vector of baseline characteristics , parallel trends hold:
Notice this is effectively a selection-on-observables assumption, but instead of , we are looking at the differenced data and arguing the mph{change} in untreated is independent of conditional on .
Then, what the Callaway and Sant’Anna estimator is combine inverse-probability of treatment weighting with a regression adjustment (a so called `doubly-robust’ method). The regression adjustment fits a model using the untreated units :
which predicts changes in given . I denote and with to emphasize this is a model for . This model can then predict the counterfactaul change in for the treated units by plugging in the treated unit’s :
This is then compared to the observed to form the DID estimate.
For estimating subgroup average effects, we can modify the procedure to take averages of
for different subgroups defined by .
This procedure is valid if is one of the covariates you use in
your conditional parallel trends assumption (passed in xformla). If
you do not include in , then you are assuming that trends do
not vary by which can be a strong assumption (do trends vary
by large / smaller counties).
Coding implementation using pte
We will use a dataset from the did package of teen employment in 500
counties in the U.S. from 2004 to 2007. The main outcome variable is
lemp which is the log of teen employment and the we will look if
minimum wage changes have larger or smaller effects in small counties
(based on lpop). First, let’s load packages and the dataset.
library(did)
library(pte) # devtools::install_github("bcallaway11/pte")
library(BMisc) # devtools::install_github("bcallaway11/BMisc")
library(fixest)
# Load example minimum wage dataset
data(mpdta, package = "did")
# Discretize population into above and below median
mpdta$large_county <- mpdta$lpop >= quantile(mpdta$lpop, 0.50)
head(mpdta)
year countyreal lpop lemp first.treat treat large_county
866 2003 8001 5.896761 8.461469 2007 1 TRUE
841 2004 8001 5.896761 8.336870 2007 1 TRUE
842 2005 8001 5.896761 8.340217 2007 1 TRUE
819 2006 8001 5.896761 8.378161 2007 1 TRUE
827 2007 8001 5.896761 8.487352 2007 1 TRUE
937 2003 8019 2.232377 4.997212 2007 1 FALSE
The main thing that pte requires of the user is a function to take a
dataframe for a given and produce a treatment effect estimate.
The dataset is effectively your original dataframe subsetted with a few
additional variables. To peek under the hood, this is what the data
looks like
# NOTE: you do not need to do this, but it's helpful to have an example to test your estimation function on
pte_data <- pte::setup_pte(
yname = "lemp", gname = "first.treat", tname = "year", idname = "countyreal", data = mpdta
)$data
# g = 2004, t = 2007
gt_data <- two_by_two_subset(pte_data, g = 2, tp = 5)$gt_data
head(gt_data)
year countyreal lpop lemp first.treat treat large_county G id
2203 2003 13011 2.668755 5.273000 0 0 FALSE 0 13011
2207 2007 13011 2.668755 5.572154 0 0 FALSE 0 13011
2210 2003 13013 3.831767 6.188264 0 0 TRUE 0 13013
2214 2007 13013 3.831767 6.401917 0 0 TRUE 0 13013
2231 2003 13019 2.787169 5.564520 0 0 FALSE 0 13019
2235 2007 13019 2.787169 4.997212 0 0 FALSE 0 13019
Y .w period name D
2203 5.273000 1 1 pre 0
2207 5.572154 1 5 post 0
2210 6.188264 1 1 pre 0
2214 6.401917 1 5 post 0
2231 5.564520 1 1 pre 0
2235 4.997212 1 5 post 0
Note we have G, id, Y (outcome), .w (weights if provided),
period (shifted version of original time), name (pre/post), and D
(treatment).
Let’s define our function. This accepts gt_data, the xformla passed,
and a custom argument called subgroup which lets you pass an
expression to subset the treated group (e.g. large_county == TRUE)
#' Subgroup ATT(g,t) estimation function
#'
#' Leave `subgroup` unspecified (or `NULL` to estimate overall ATT(g,t)
#' Otherwise pass expression to `subgroup` to subset the treated units to a subgroup
#'
did_attgt_subgroup <- function(gt_data, xformla, subgroup = NULL, ...) {
# convert two period panel into wide with `pre` and `post`
data_pre <- gt_data[gt_data$name == "pre", ]
names(data_pre)[names(data_pre) == "Y"] <- "Y_pre"
data_post <- gt_data[gt_data$name == "post", ]
names(data_post)[names(data_post) == "Y"] <- "Y_post"
data_post <- data_post[, c("id", "Y_post")]
# One row per unit
wide_data <- merge(data_pre, data_post, by = "id")
# \Delta Y = Y_{i,t} - Y_{i,g-1}
wide_data$delta_Y <- wide_data$Y_post - wide_data$Y_pre
# Fit model using D == 0
outcome_reg <- fixest::feols(
fixest::xpd(lhs = "delta_Y", fml = xformla),
data = wide_data[wide_data$D == 0, ] # never-treated and/or not-yet-treated
)
# Predict Y_{it}(0) - Y_{i,g-1}(0) for everyone
wide_data$delta_Y0_hat <- predict(outcome_reg, newdata = wide_data)
# Difference between observed delta_y and predicted delta_Y0_hat
wide_data$diff <- wide_data$delta_Y - wide_data$delta_Y0_hat
# Get treated units
treat <- wide_data[wide_data$D == 1, ]
# Using a little quosure magic to allow `subgroup` to be passed in as an argument
subgroup <- rlang::enquo(subgroup)
if (!rlang::quo_is_null(subgroup)) {
subgroup <- rlang::eval_tidy(subgroup, data = treat)
treat <- treat[subgroup, ]
}
# Estimate ATT for treated units
attgt <- mean(treat$diff)
return(list(attgt = attgt))
}
Now, with that function defined we can use it with pte. The function
takes very similar arguments to did (variable names and xformla),
but requires a few extra arguments (setup_pte_fun and subset_fun)
given the extra flexibility given by the pte package.
# Overall ATT
did_res <- pte(
data = mpdta,
yname = "lemp",
gname = "first.treat",
tname = "year",
idname = "countyreal",
xformla = ~lpop,
setup_pte_fun = pte::setup_pte,
subset_fun = pte::two_by_two_subset,
# base_period = "universal",
biters = 50,
# No subgroup filter so it estimates an overal ATT(g,t)
attgt_fun = did_attgt_subgroup
)
summary(did_res)
Overall ATT:
ATT Std. Error [ 95% Conf. Int.]
-0.0323 0.0127 -0.0572 -0.0073 *
Dynamic Effects:
Event Time Estimate Std. Error [95% Pointwise Conf. Band]
-3 0.0267 0.0148 -0.0023 0.0558
-2 -0.0051 0.0116 -0.0279 0.0176
-1 -0.0233 0.0145 -0.0517 0.0051
0 -0.0200 0.0125 -0.0446 0.0046
1 -0.0550 0.0115 -0.0776 -0.0323 *
2 -0.1385 0.0357 -0.2085 -0.0685 *
3 -0.1075 0.0340 -0.1742 -0.0409 *
---
Signif. codes: `*' confidence band does not cover 0
# Check similar results using `did` when using Outcome Regression
did::aggte(
did::att_gt(
data = mpdta,
yname = "lemp",
gname = "first.treat",
tname = "year",
idname = "countyreal",
xformla = ~lpop,
est_method = "reg"
),
"dynamic"
)
Call:
did::aggte(MP = did::att_gt(data = mpdta, yname = "lemp", gname = "first.treat",
tname = "year", idname = "countyreal", xformla = ~lpop, est_method = "reg"),
type = "dynamic")
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
Overall summary of ATT's based on event-study/dynamic aggregation:
ATT Std. Error [ 95% Conf. Int.]
-0.0808 0.0187 -0.1174 -0.0441 *
Dynamic Effects:
Event time Estimate Std. Error [95% Simult. Conf. Band]
-3 0.0264 0.0142 -0.0097 0.0624
-2 -0.0041 0.0130 -0.0371 0.0288
-1 -0.0235 0.0146 -0.0607 0.0138
0 -0.0211 0.0117 -0.0509 0.0086
1 -0.0534 0.0163 -0.0949 -0.0118 *
2 -0.1411 0.0394 -0.2414 -0.0408 *
3 -0.1075 0.0332 -0.1921 -0.0230 *
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Outcome Regression
Now, we can pass the subgroup argument to estimate effects for large
counties and for small counties separately.
did_res_Z0 <- pte(
data = mpdta,
yname = "lemp",
gname = "first.treat",
tname = "year",
idname = "countyreal",
xformla = ~lpop,
setup_pte_fun = pte::setup_pte,
subset_fun = pte::two_by_two_subset,
# base_period = "universal",
biters = 50,
attgt_fun = did_attgt_subgroup,
subgroup = (large_county == FALSE)
)
summary(did_res_Z0)
Overall ATT:
ATT Std. Error [ 95% Conf. Int.]
-0.0301 0.0233 -0.0757 0.0154
Dynamic Effects:
Event Time Estimate Std. Error [95% Pointwise Conf. Band]
-3 0.0284 0.0237 -0.0180 0.0748
-2 -0.0148 0.0199 -0.0539 0.0243
-1 -0.0249 0.0268 -0.0774 0.0277
0 -0.0149 0.0222 -0.0583 0.0286
1 -0.0902 0.0358 -0.1605 -0.0199 *
2 -0.2100 0.0534 -0.3147 -0.1054 *
3 -0.1413 0.0575 -0.2541 -0.0286 *
---
Signif. codes: `*' confidence band does not cover 0
did_res_Z1 <- pte(
data = mpdta,
yname = "lemp",
gname = "first.treat",
tname = "year",
idname = "countyreal",
xformla = ~lpop,
setup_pte_fun = pte::setup_pte,
subset_fun = pte::two_by_two_subset,
# base_period = "universal",
biters = 50,
attgt_fun = did_attgt_subgroup,
subgroup = (large_county == TRUE)
)
summary(did_res_Z1)
Overall ATT:
ATT Std. Error [ 95% Conf. Int.]
-0.0338 0.0086 -0.0508 -0.0169 *
Dynamic Effects:
Event Time Estimate Std. Error [95% Pointwise Conf. Band]
-3 0.0255 0.0112 0.0035 0.0474 *
-2 0.0012 0.0103 -0.0190 0.0213
-1 -0.0216 0.0183 -0.0574 0.0142
0 -0.0247 0.0085 -0.0413 -0.0081 *
1 -0.0275 0.0198 -0.0662 0.0113
2 -0.0669 0.0203 -0.1066 -0.0272 *
3 -0.0738 0.0234 -0.1196 -0.0279 *
---
Signif. codes: `*' confidence band does not cover 0
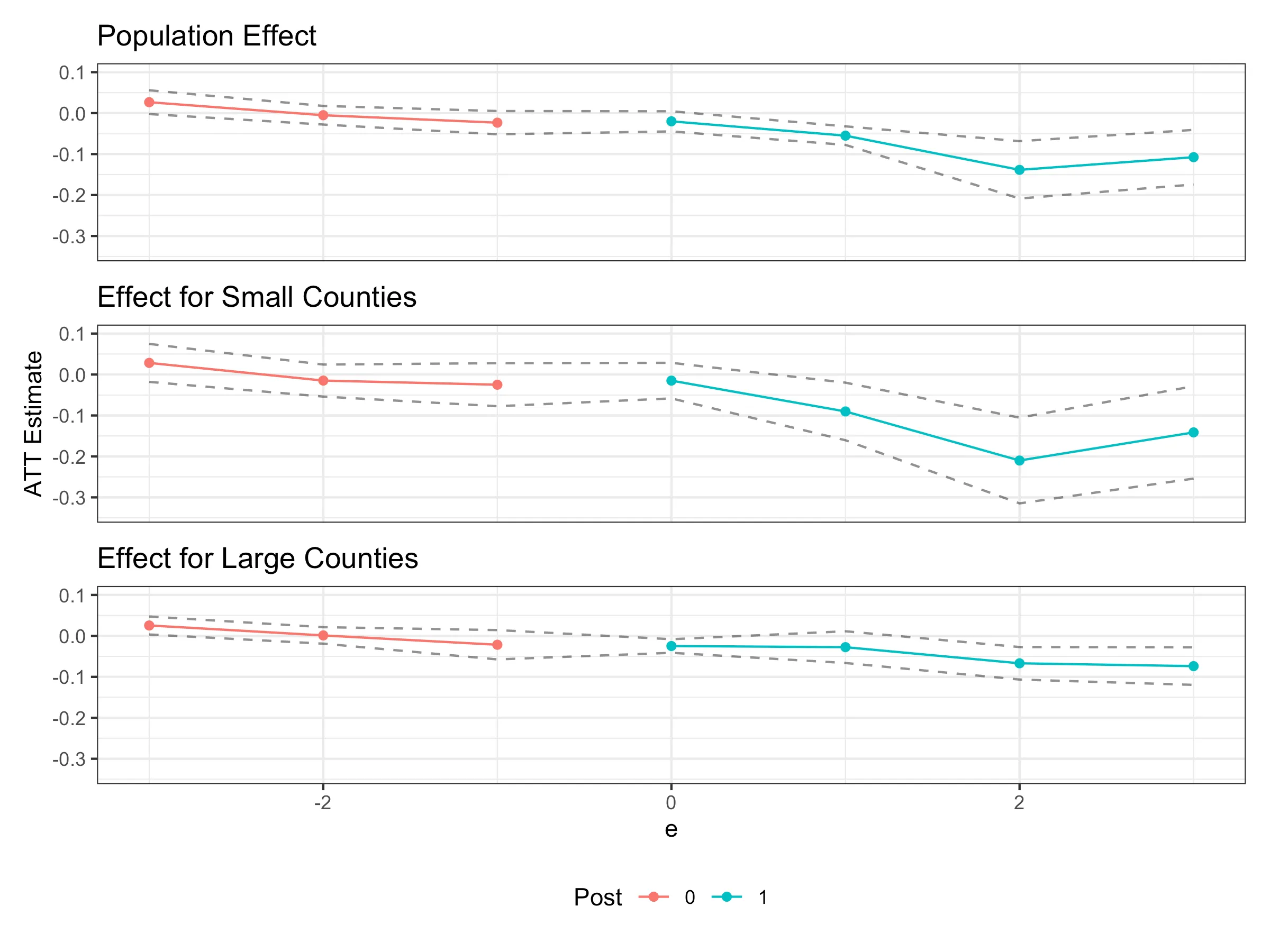
Plotting results
library(ggplot2)
library(patchwork)
plot_overall <- pte:::ggpte(did_res) +
labs(title = "Population Effect") +
theme(legend.position = "bottom")
plot_Z0 <- pte:::ggpte(did_res_Z0) +
labs(title = "Effect for Small Counties") +
theme(legend.position = "bottom")
plot_Z1 <- pte:::ggpte(did_res_Z1) +
labs(title = "Effect for Large Counties") +
theme(legend.position = "bottom")
# Combine figures into one plot
((plot_overall / plot_Z0 / plot_Z1) &
scale_y_continuous(limits = c(-0.34, 0.1)) &
labs(y = "ATT Estimate", color = "Post") &
theme(legend.position = "bottom")
) +
plot_layout(axes = "collect", guides = "collect")
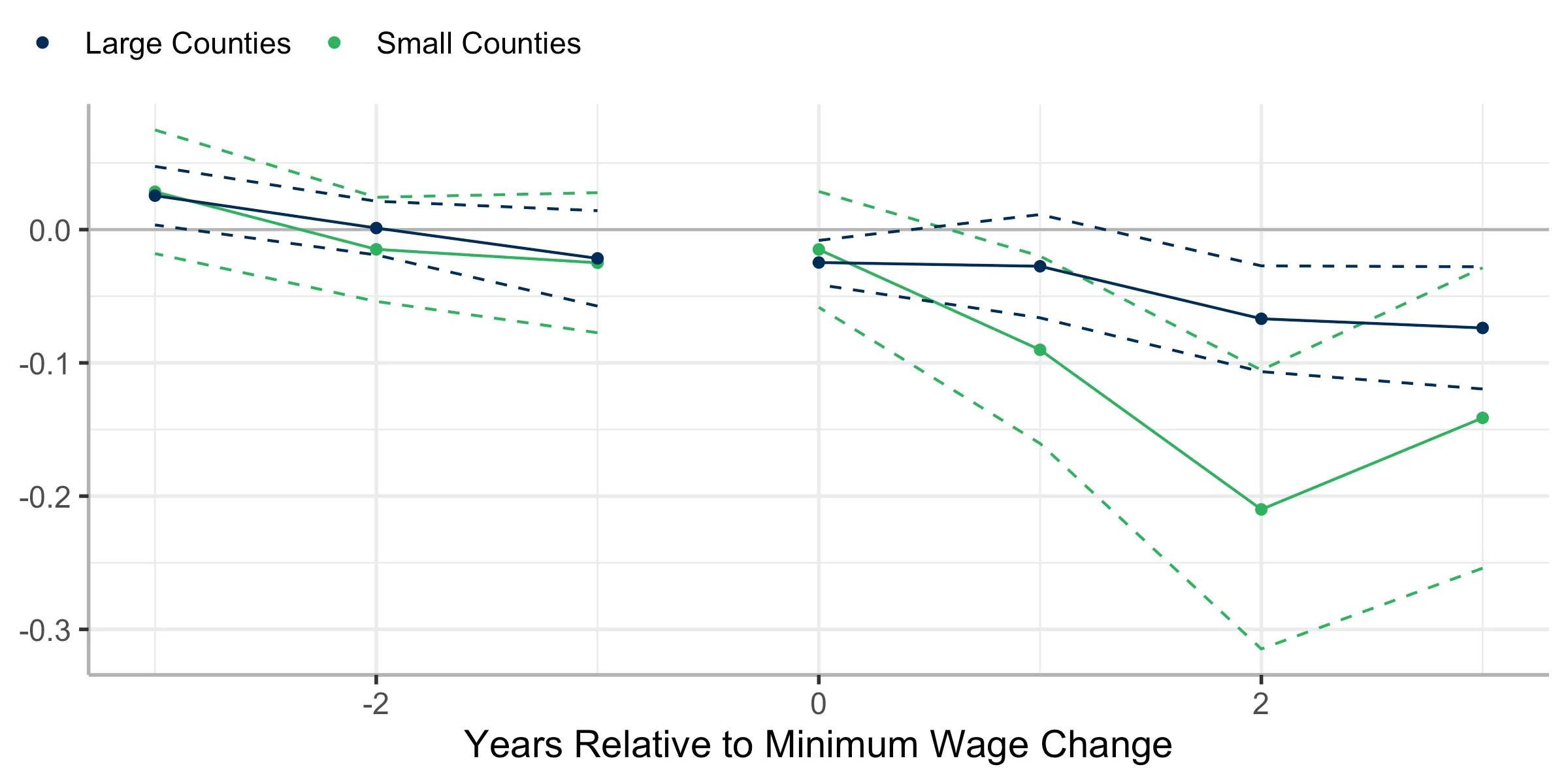
Or, you can extract the event-study estimates directly using summary()
and plot the estimates however you wish.
plot_df_Z0 <- summary(did_res_Z0)$event_study
colnames(plot_df_Z0) <- c("e", "att", "se", "cil", "ciu")
plot_df_Z1 <- summary(did_res_Z1)$event_study
colnames(plot_df_Z1) <- c("e", "att", "se", "cil", "ciu")
ggplot() +
geom_hline(yintercept = 0, color = "grey70") +
geom_line(
aes(x = e, y = att, color = "Small Counties", group = (e >= 0)),
data = plot_df_Z0
) +
geom_point(
aes(x = e, y = att, color = "Small Counties"),
data = plot_df_Z0
) +
geom_ribbon(
aes(x = e, ymin = cil, ymax = ciu, color = "Small Counties", group = (e >= 0)),
data = plot_df_Z0,
fill = NA, linetype = "dashed"
) +
geom_line(
aes(x = e, y = att, color = "Large Counties", group = (e >= 0)),
data = plot_df_Z1
) +
geom_point(
aes(x = e, y = att, color = "Large Counties"),
data = plot_df_Z1
) +
geom_ribbon(
aes(x = e, ymin = cil, ymax = ciu, color = "Large Counties", group = (e >= 0)),
data = plot_df_Z1,
fill = NA, linetype = "dashed"
) +
labs(x = "Years Relative to Minimum Wage Change", y = NULL, color = NULL) +
scale_color_manual(
values = c(
"Large Counties" = "#002C55",
"Small Counties" = "#2DB25F"
),
guide = guide_legend(override.aes = list(linetype = "blank"))
) +
theme_bw(base_size = 14) +
theme(
panel.border = element_blank(), axis.line = element_line(color = "grey70"),
legend.position = "top",
legend.margin = margin(0, 0, 0, 0),
legend.justification = c(0, 1),
legend.location = "plot"
)
References
Citation:
@online{callaway-unconfoundedness-regressions-2024, author = {Callaway, Brantly}, title = {Interpreting Regressions under the Assumption of Unconfoundedness}, year = {2024}, month = {11}, url = {https://bcallaway11.github.io/posts/unconfoundedness-regressions}, urldate = {\today}, }